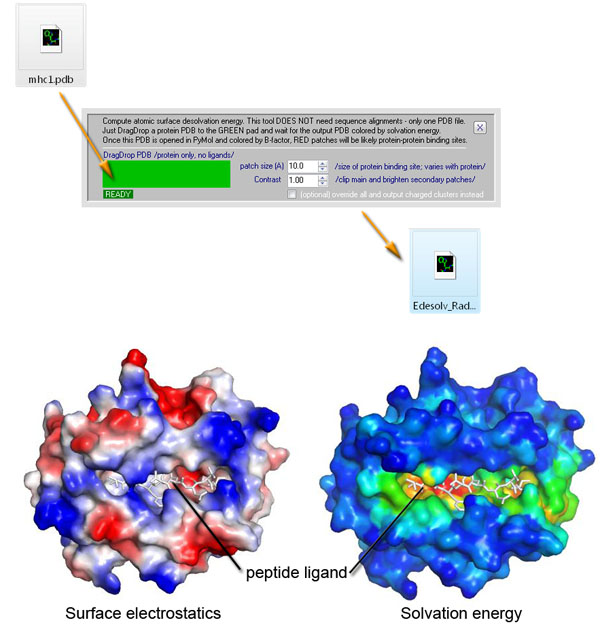
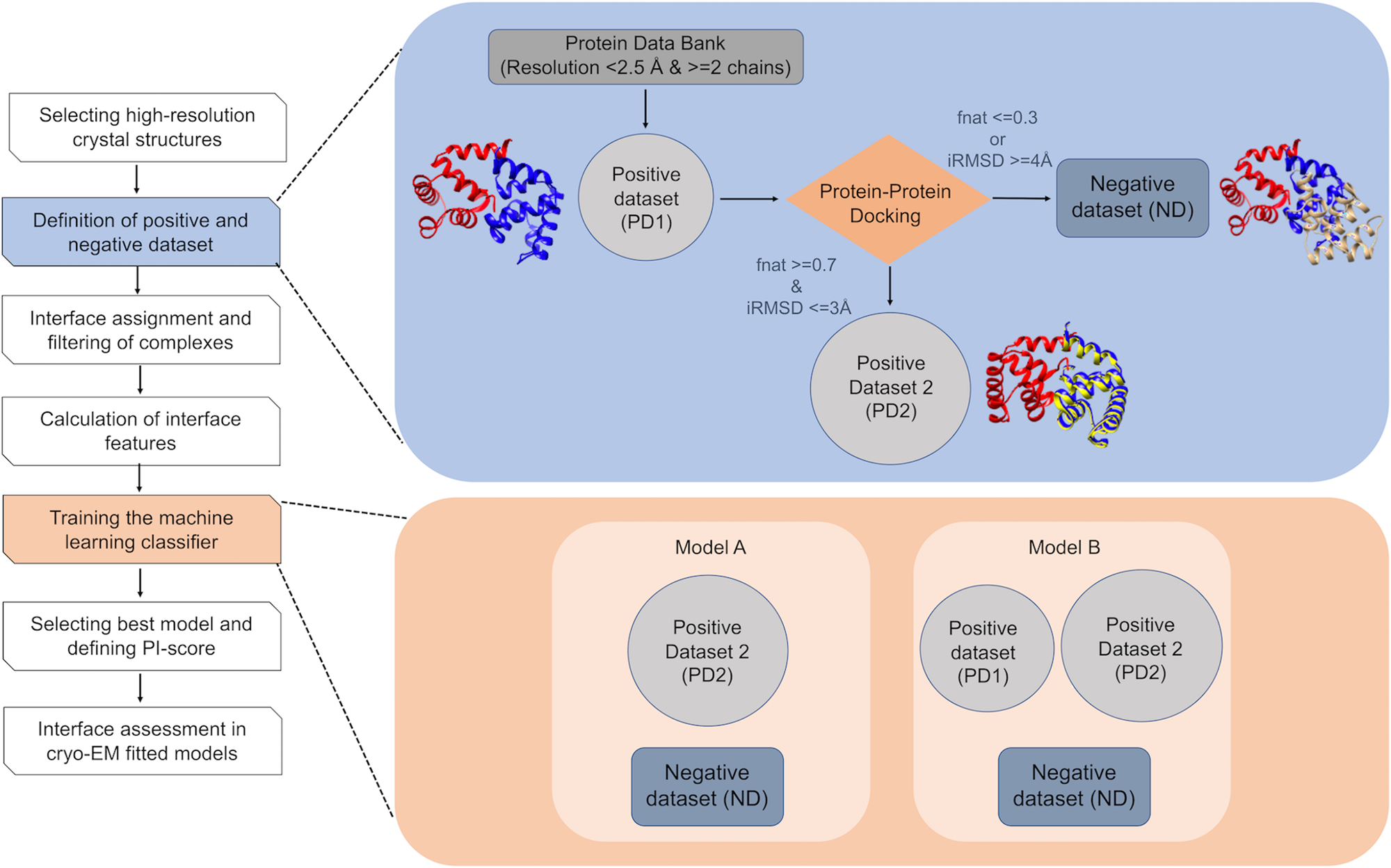
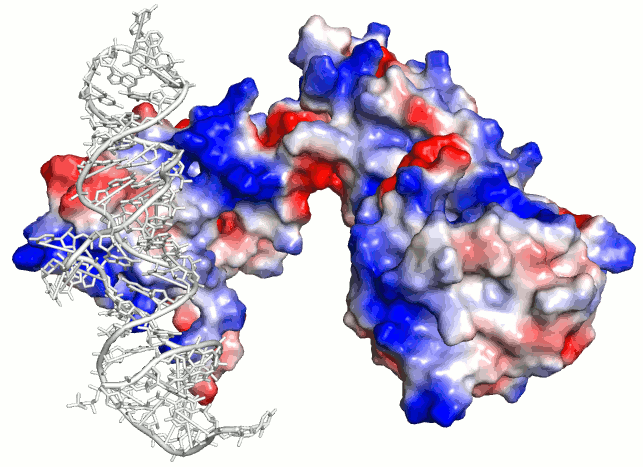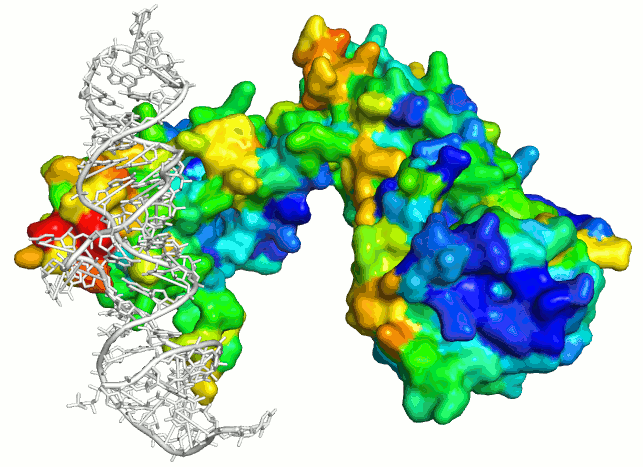
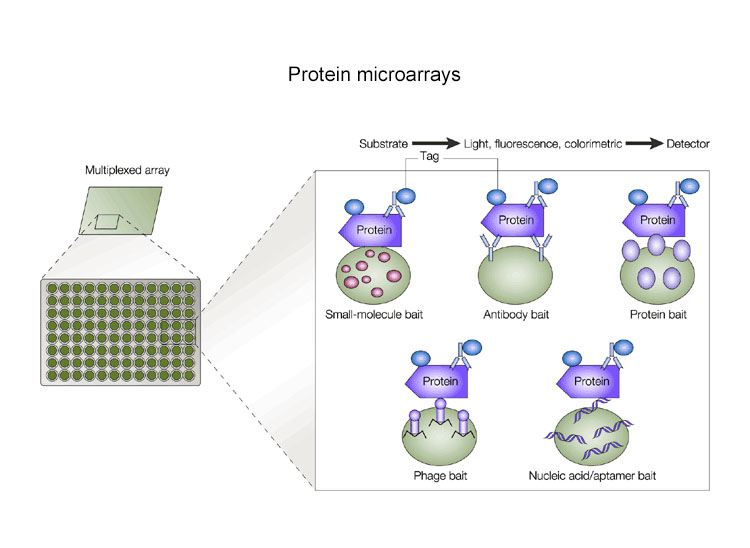
Predicting Protein–Protein Interfaces that Bind Intrinsically Disordered Protein Regions - ScienceDirect
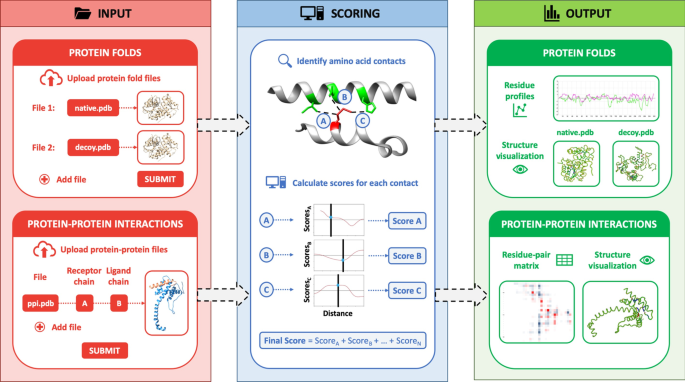
SPServer: split-statistical potentials for the analysis of protein structures and protein–protein interactions | BMC Bioinformatics | Full Text

Recent advances in predicting and modeling protein–protein interactions: Trends in Biochemical Sciences

Identifying protein-protein interface via a novel multi-scale local sequence and structural representation | BMC Bioinformatics | Full Text

AREA-AFFINITY: A Web Server for Machine Learning-Based Prediction of Protein –Protein and Antibody–Protein Antigen Binding Affinities | Journal of Chemical Information and Modeling

BindML: Software for Predicting Protein-Protein Interaction Sites using Phylogenetic Substitution Models
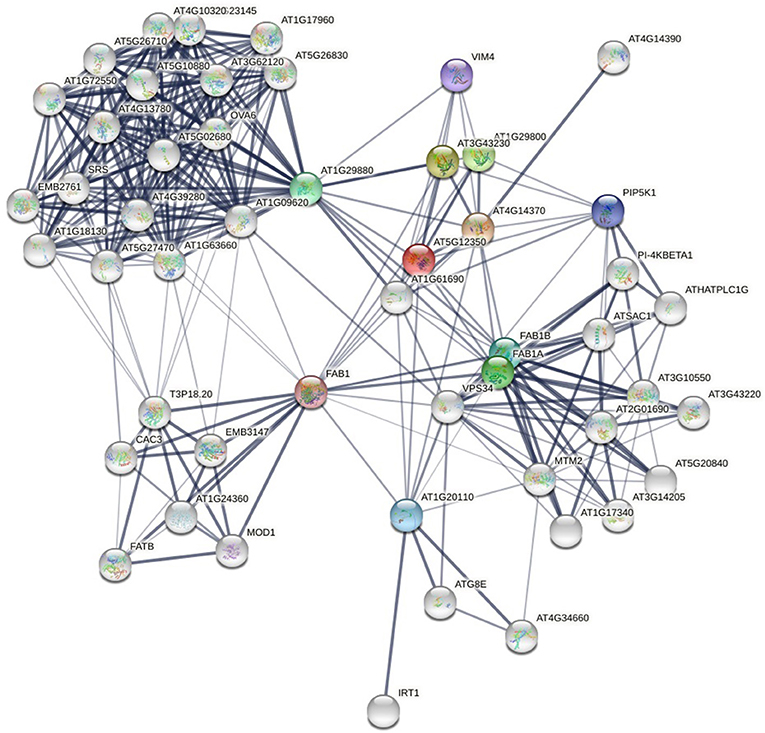
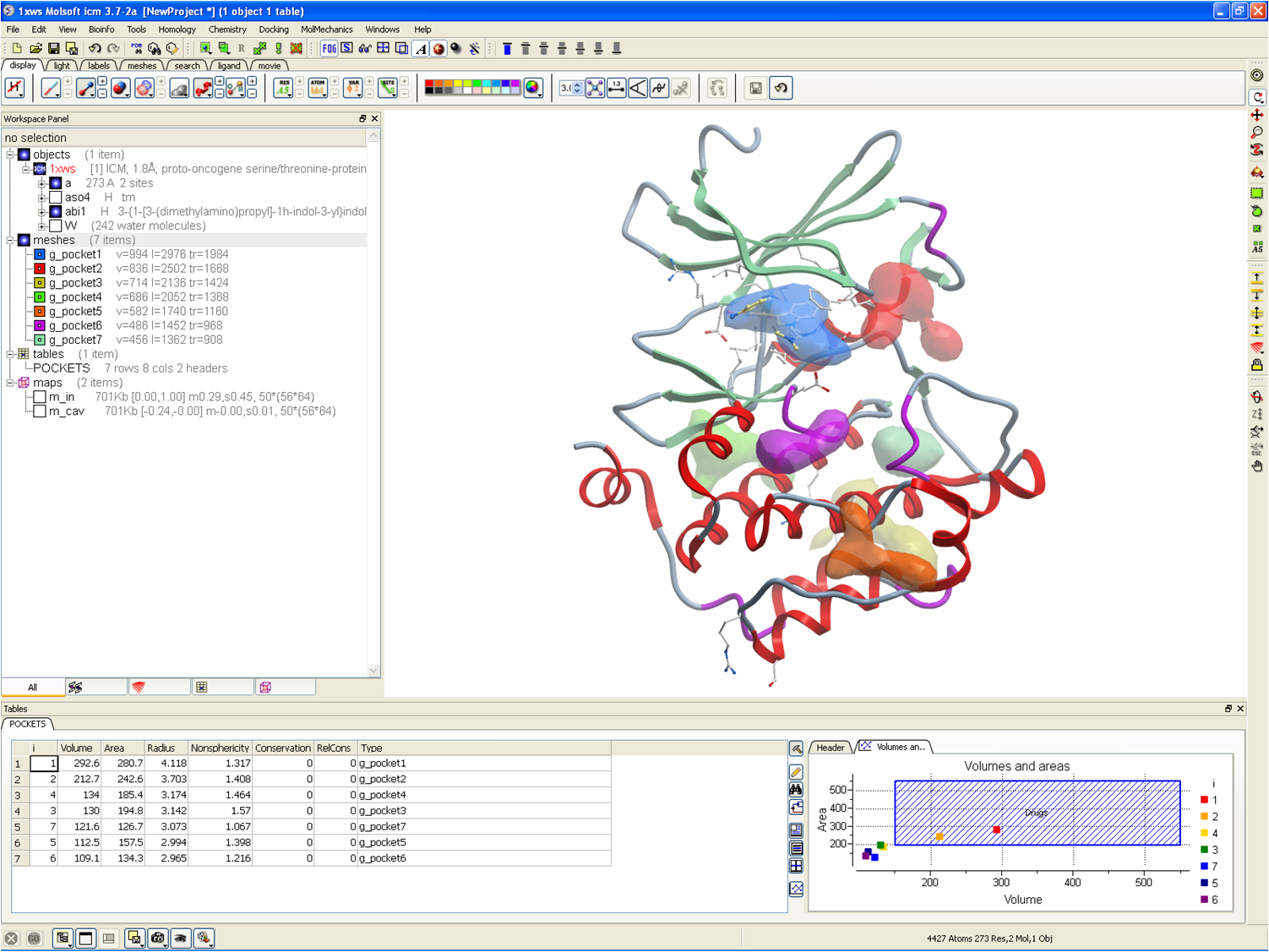
Frontiers | Protein Interface Complementarity and Gene Duplication Improve Link Prediction of Protein-Protein Interaction Network

Arpeggio: A Web Server for Calculating and Visualising Interatomic Interactions in Protein Structures | Semantic Scholar
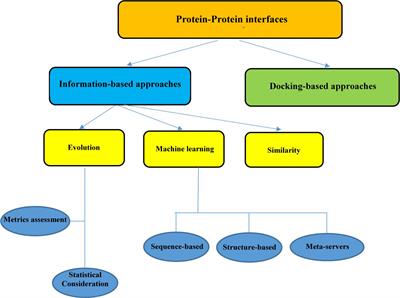
Frontiers | In silico Approaches for the Design and Optimization of Interfering Peptides Against Protein–Protein Interactions

Uncovering the basis of protein-protein interaction specificity with a combinatorially complete library | eLife

Large-scale discovery of protein interactions at residue resolution using co-evolution calculated from genomic sequences | Nature Communications

Classification and prediction of protein–protein interaction interface using machine learning algorithm | Scientific Reports

Finding the ΔΔG spot: Are predictors of binding affinity changes upon mutations in protein–protein interactions ready for it? - Geng - 2019 - WIREs Computational Molecular Science - Wiley Online Library

Computational prediction of protein–protein binding affinities - Siebenmorgen - 2020 - WIREs Computational Molecular Science - Wiley Online Library

Binding mode of each peptide-protein complex using Protein Interaction... | Download Scientific Diagram